Visualization for diagnostic review of copy number variants in complex DNA sequencing data
Emilia Ståhlbom -
Jesper Molin -
Claes Lundström -
Anders Ynnerman -
DOI: 10.1109/TVCG.2024.3385118
Room: Bayshore I
2024-10-16T14:51:00ZGMT-0600Change your timezone on the schedule page
2024-10-16T14:51:00Z
Fast forward
Full Video
Keywords
Visualization, genomics, copy number variants, clinical decision support, evaluation
Abstract
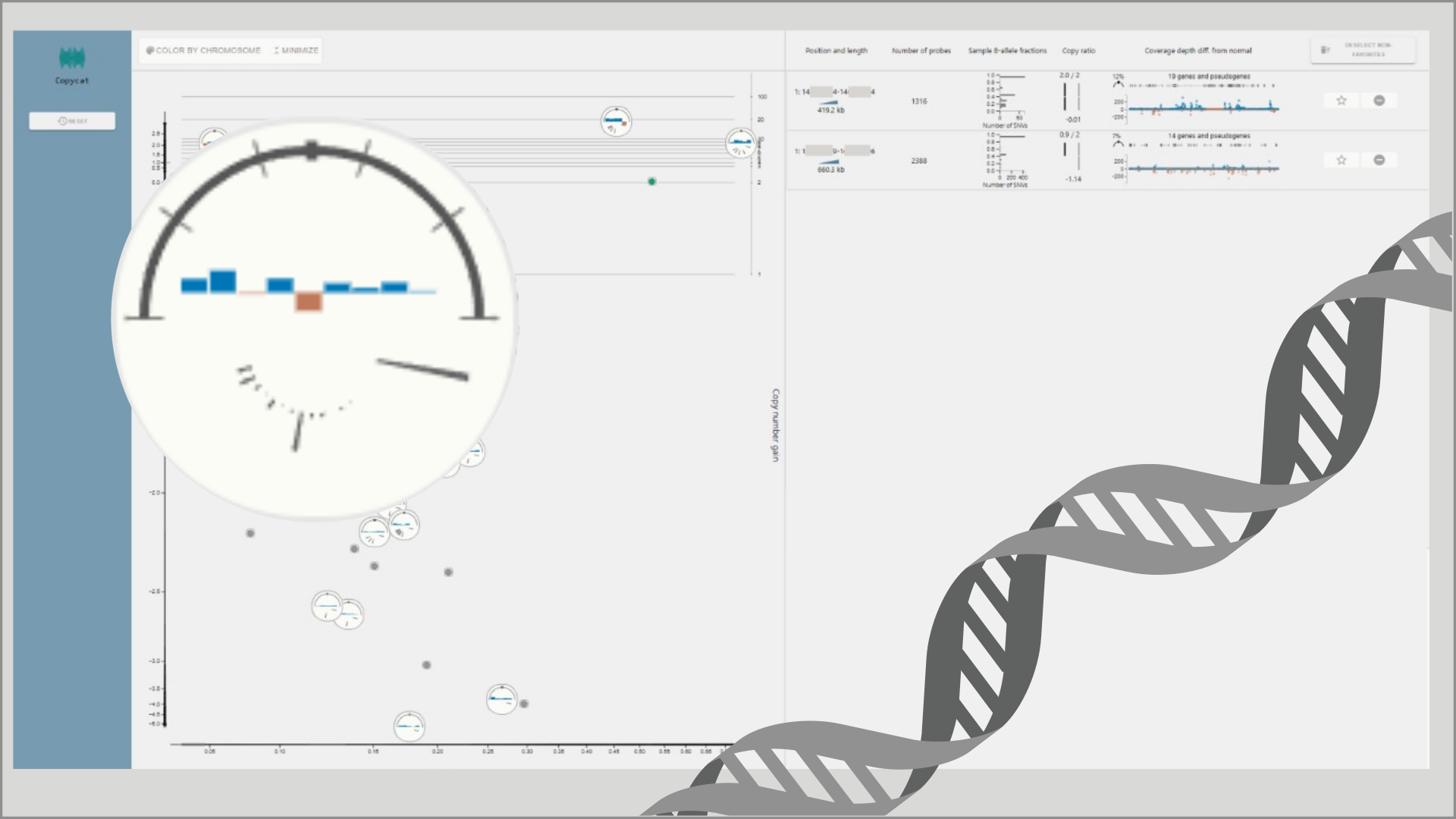
Genomics is at the core of precision medicine, and there are high expectations on genomics-enabled improvement of patient outcomes in the years to come. Around the world, initiatives to increase the use of DNA sequencing in clinical routine are being deployed, such as the use of broad panels in the standard care for oncology patients. Such a development comes at the cost of increased demands on throughput in genomic data analysis. In this paper, we use the task of copy number variant (CNV) analysis as a context for exploring visualization concepts for clinical genomics. CNV calls are generated algorithmically, but time-consuming manual intervention is needed to separate relevant findings from irrelevant ones in the resulting large call candidate lists. We present a visualization environment, named Copycat, to support this review task in a clinical scenario.Key components are a scatter-glyph plot replacing the traditional list visualization, and a glyph representation designed for at-a-glance relevance assessments. Moreover, we present results from a formative evaluation of the prototype by domain specialists, from which we elicit insights to guide both prototype improvements and visualization for clinical genomics in general.