A General Framework for Comparing Embedding Visualizations Across Class-Label Hierarchies
Trevor Manz - Harvard Medical School, Boston, United States
Fritz Lekschas - Ozette Technologies, Seattle, United States
Evan Greene - Ozette Technologies, Seattle, United States
Greg Finak - Ozette Technologies, Seattle, United States
Nils Gehlenborg - Harvard Medical School, Boston, United States
Screen-reader Accessible PDF
Download preprint PDF
Room: Bayshore I
2024-10-17T12:42:00ZGMT-0600Change your timezone on the schedule page
2024-10-17T12:42:00Z
Fast forward
Full Video
Keywords
visualization, comparison, high-dimensional data, dimensionality reduction, embeddings
Abstract
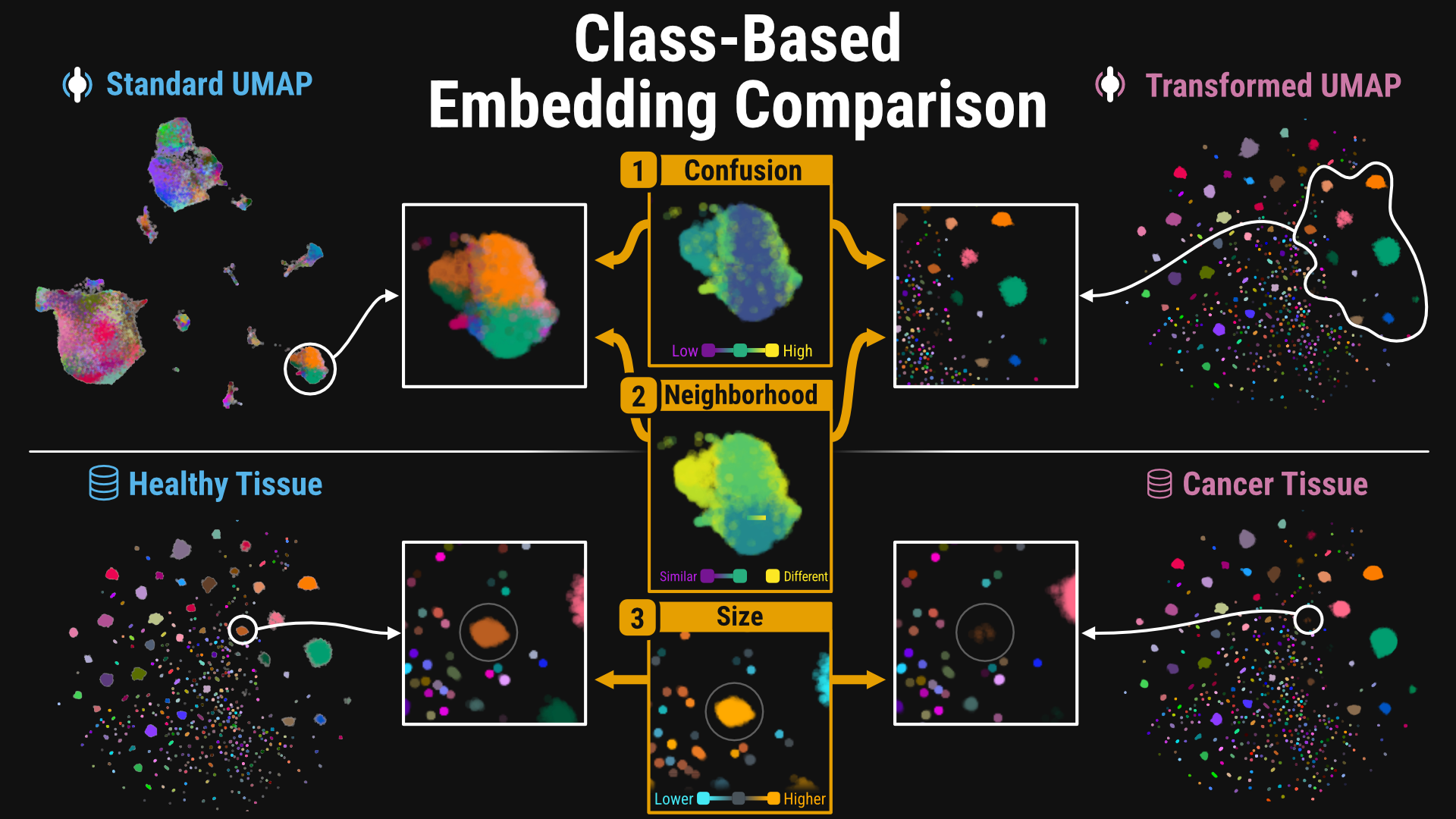
Projecting high-dimensional vectors into two dimensions for visualization, known as embedding visualization, facilitates perceptual reasoning and interpretation. Comparing multiple embedding visualizations drives decision-making in many domains, but traditional comparison methods are limited by a reliance on direct point correspondences. This requirement precludes comparisons without point correspondences, such as two different datasets of annotated images, and fails to capture meaningful higher-level relationships among point groups. To address these shortcomings, we propose a general framework for comparing embedding visualizations based on shared class labels rather than individual points. Our approach partitions points into regions corresponding to three key class concepts—confusion, neighborhood, and relative size—to characterize intra- and inter-class relationships. Informed by a preliminary user study, we implemented our framework using perceptual neighborhood graphs to define these regions and introduced metrics to quantify each concept.We demonstrate the generality of our framework with usage scenarios from machine learning and single-cell biology, highlighting our metrics' ability to draw insightful comparisons across label hierarchies. To assess the effectiveness of our approach, we conducted an evaluation study with five machine learning researchers and six single-cell biologists using an interactive and scalable prototype built with Python, JavaScript, and Rust. Our metrics enable more structured comparisons through visual guidance and increased participants’ confidence in their findings.