Best Paper Award
Aardvark: Composite Visualizations of Trees, Time-Series, and Images
Devin Lange - University of Utah, Salt Lake City, United States
Robert L Judson-Torres - University of Utah, Salt Lake City, United States
Thomas A Zangle - University of Utah, Salt Lake City, United States
Alexander Lex - University of Utah, Salt Lake City, United States
Screen-reader Accessible PDF
Download preprint PDF
Download Supplemental Material
Room: Bayshore I + II + III
2024-10-15T16:25:00ZGMT-0600Change your timezone on the schedule page
2024-10-15T16:25:00Z
Fast forward
Full Video
Keywords
Visualization, Cell Microscopy, View Composition
Abstract
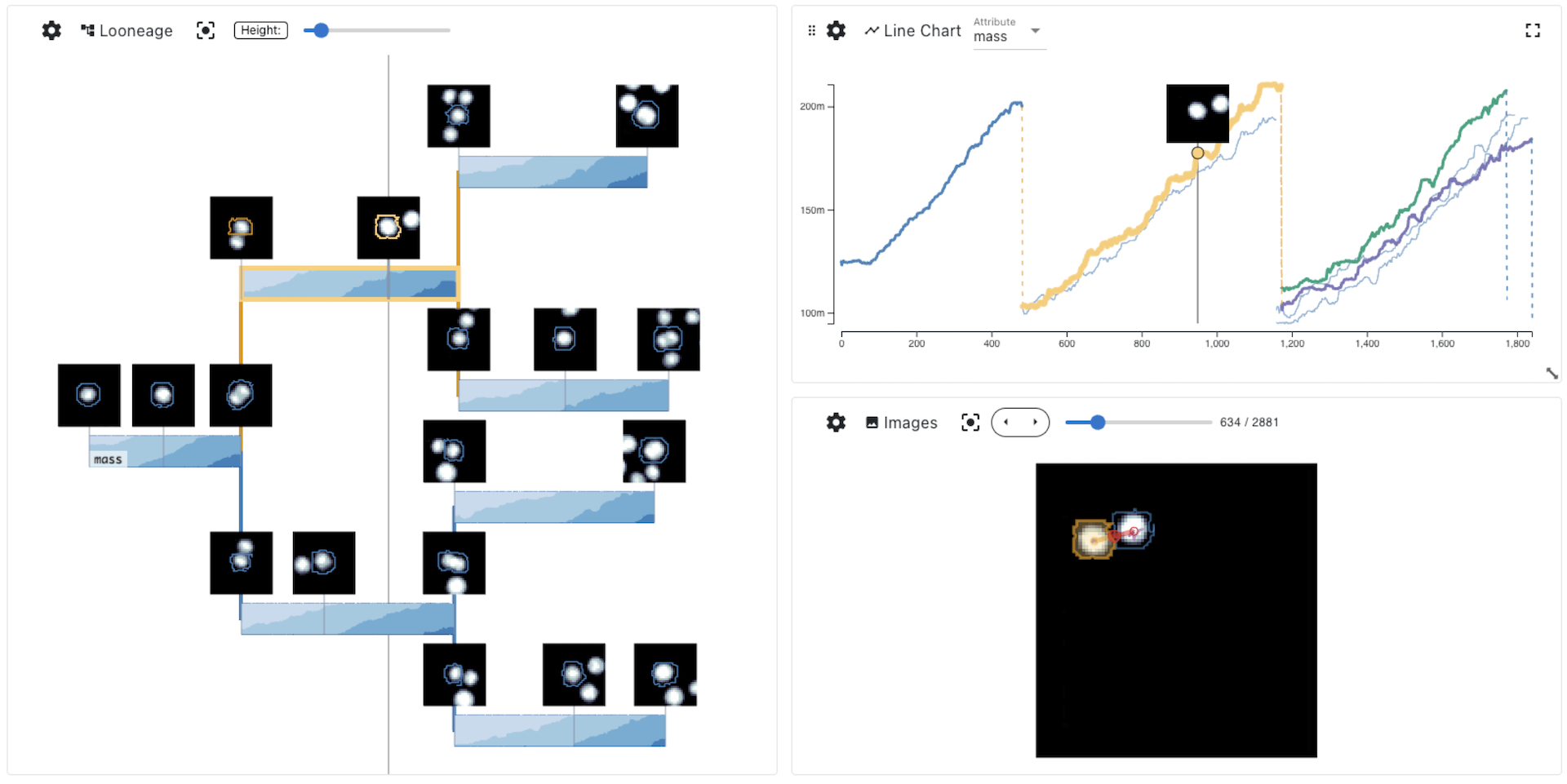
How do cancer cells grow, divide, proliferate, and die? How do drugs influence these processes? These are difficult questions that we can attempt to answer with a combination of time-series microscopy experiments, classification algorithms, and data visualization.However, collecting this type of data and applying algorithms to segment and track cells and construct lineages of proliferation is error-prone; and identifying the errors can be challenging since it often requires cross-checking multiple data types. Similarly, analyzing and communicating the results necessitates synthesizing different data types into a single narrative. State-of-the-art visualization methods for such data use independent line charts, tree diagrams, and images in separate views. However, this spatial separation requires the viewer of these charts to combine the relevant pieces of data in memory. To simplify this challenging task, we describe design principles for weaving cell images, time-series data, and tree data into a cohesive visualization. Our design principles are based on choosing a primary data type that drives the layout and integrates the other data types into that layout. We then introduce Aardvark, a system that uses these principles to implement novel visualization techniques. Based on Aardvark, we demonstrate the utility of each of these approaches for discovery, communication, and data debugging in a series of case studies.